Click in the image to view a video
ZEBRA is an embryology dataset
The ZEBRA dataset provides a feature representation (154
features) of cells of zebrafish embryo to determine whether they are in
division (meiosis) or not. All the examples are manually annotated. The
dataset was prepared by Emmanuel
Faure (Institut
des systèmes complexes, France).
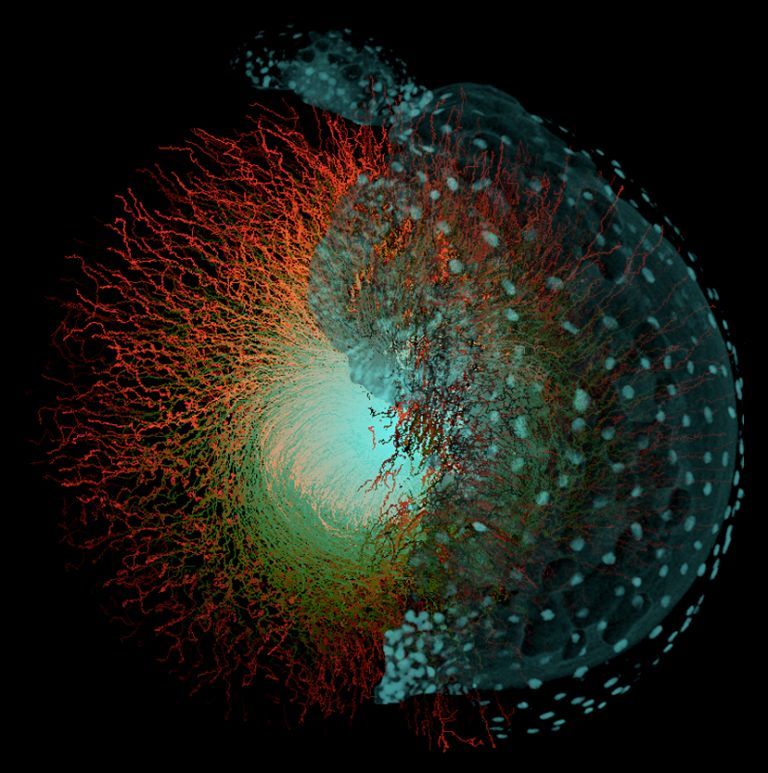
The embryomics project is
devoted to the morphodynamical “reconstruction” of the cell lineage
tree
underlying the processes of animal embryogenesis. A set of
strategies, methods and algorithms to “sequence” the cell lineage tree
as a
branching process annotated in space and time was designed. The final
goal is
to fully
reconstruct the dynamics of cell divisions and movements from
time-lapse series
of high-resolution optical sections obtained by multiphoton laser
scanning
microscopy throughout embryonic development of live animals. Embryomics
allows
the automated tracking of events such as cell division and cell death
in live
embryos and give us access to parameters such as the rate of cell
proliferation
in time and space. Special
thanks to all the
project participants: Thierry Savy, Louise Duloquin, Miguel Luengo
Oroz, Benoit Lombardot, Camilo Melani, Paul Bourgine, and Nadine
Peyriéras.